16 Case 3: Burden in Batch
Case 3: Site-Level Analysis of Non-B Burden Heterogeneity among multiple mutations and samples (Multiple groups of multiple regions)
Use cases: The goal of this use case is to demonstrate the ability to explore non-B burden localized to site-level genomic coordinates from multiple genes and samples with use of the ‘burden in batch’ option.
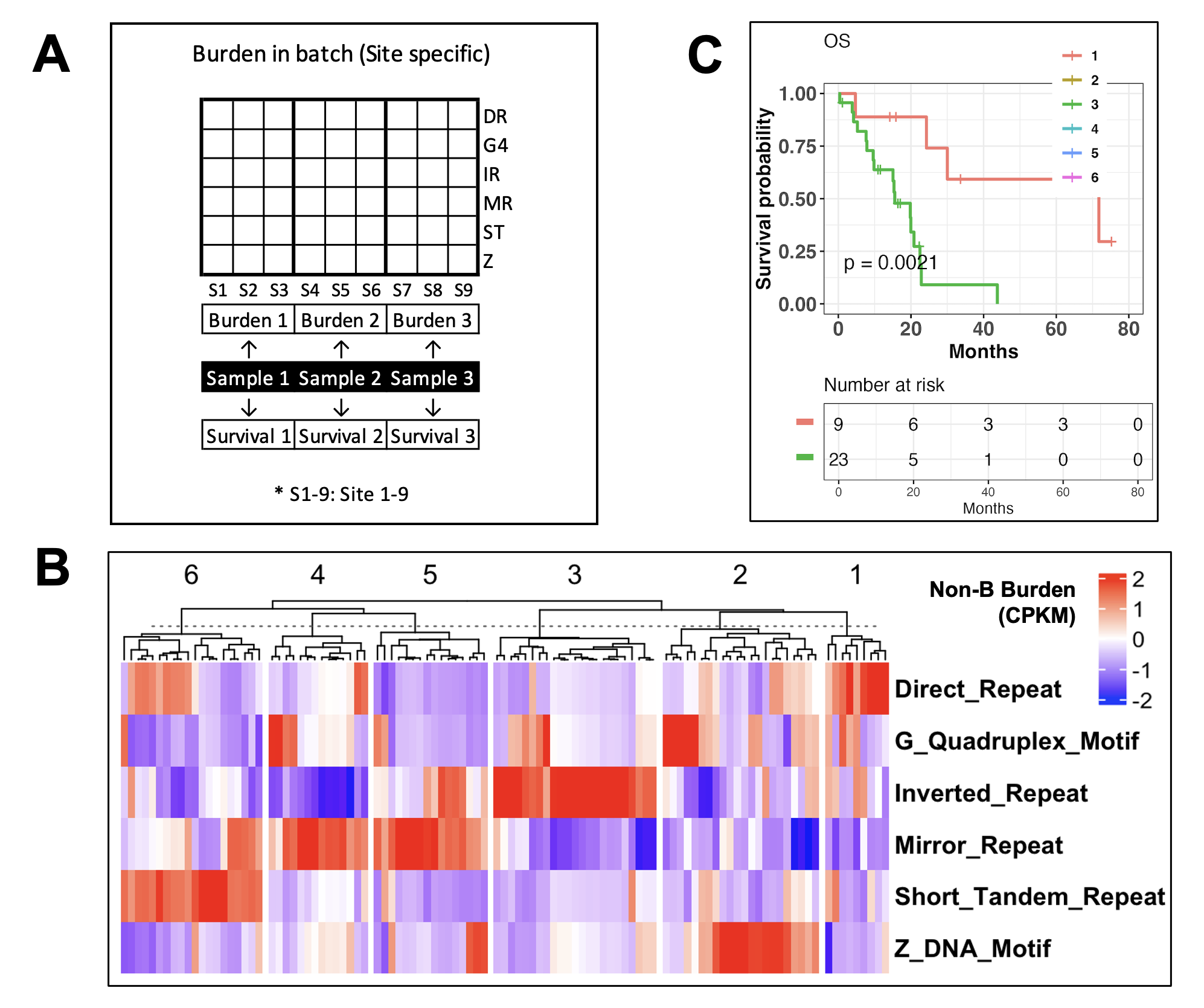
Example: We applied mutation-localized non-B burdens calculation to genome-wide mutation sites from 104 early-stage pancreatic cancer patients with mutation and survival data from TCGA. In other words, 104 groups of genomic mutation regions from 104 samples were used as input for burden in batch calculation (A). Each group has its own specific mutation sites signature per sample. The mutations sites of each group were overlapped with non-B forming motif regions to calculate the non-B burden within each sample.
Results: For each sample, we derived a site-level non-B burden for each non-B DNA structure, resulting in a non-B burden output matrix of 104 (columns, input groups) x 6 (rows, non-B types) (B). We performed a cluster analysis on these non-B burdens and compared overall survival between groups (C). Among the 104 early-stage pancreatic patients, non-B burden clustering resulted in six patient clusters that differentiated by non-B DNA structures burden, in which IR high burden samples (n=23, median OS=15 month) significantly differed in OS from DR high burden samples (n=23, median OS=30 month). The resulting output matrix of burdens on these sample can be used for other downstream analyses including supervise and unsupervised clustering, total burden calculation, association analyses and more depending on research questions.