4 Input formats
The web server provides four options with which to satisfy users’ input requirements:
The first option includes built-in cancer related signature gene sets from which the user can select that include DNA damage repair and response gene pathways, cancer hallmark gene set, oncogenes etc.
A second built-in input option for user selection includes cancer cell line-specific molecular features that include mutations and copy number alterations.
The third input option allows users to manually input a single or several genes through the web interfaces for when a quick gene query is of interest.
The fourth input option allows users to upload a set of genomic coordinates representing genomic regions of interest, such as mutation sites, in which non-B burden is placed in the context of mutation-localized non-B burden.
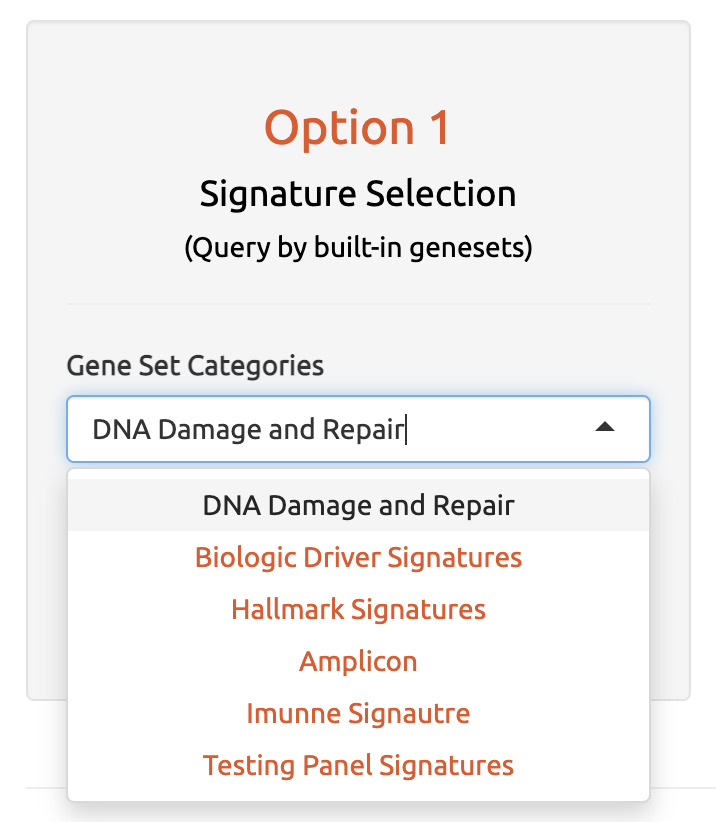
4.1 Option 1: By Cancer Gene Signatures
The first input option includes built-in cancer-related signature gene sets, such as DNA damage repair and response gene pathways, cancer hallmark gene sets, oncogenes, and more. Users can select from these pre-defined sets to perform non-B burden calculations for these specific gene sets.
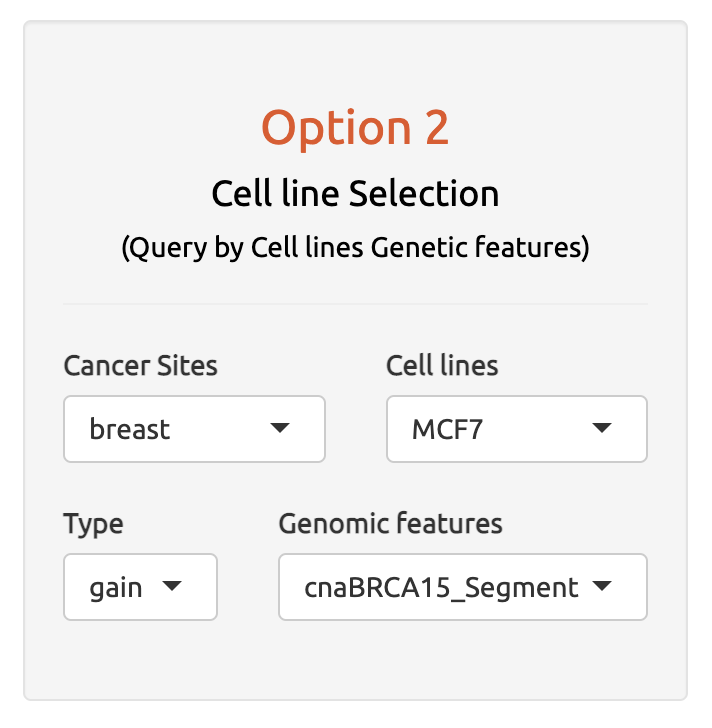
4.2 Option 2: By Cancer Cell Lines
By Cancer cell lines and molecular genomic features (SNV, CNV)
A second input option allows users to select gene sets based on specific cancer cell lines of interest and their molecular features, such as mutations and copy number alterations. This option enables users to perform non-B burden calculations for gene sets specific to the selected cancer cell lines.
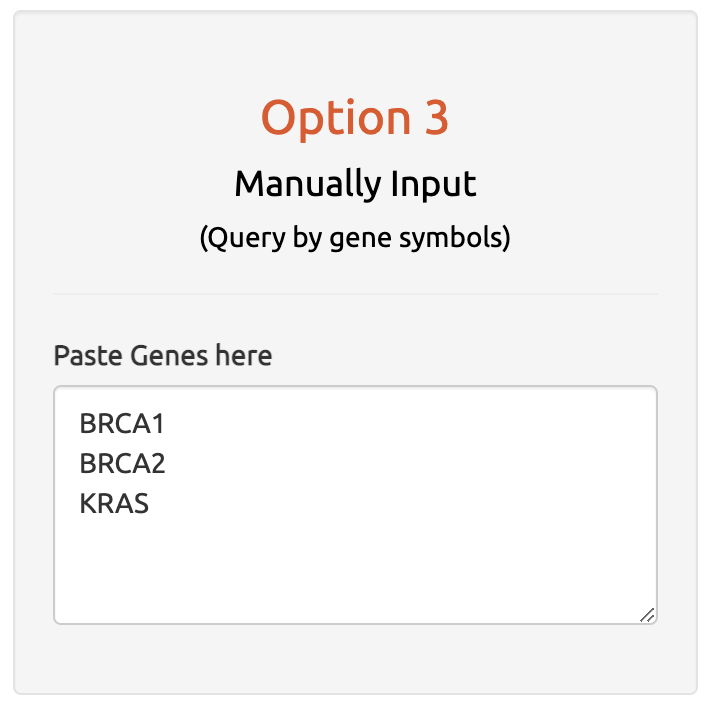
4.3 Option 3: Manually Input
Manually input gene symbol for user-defined input
The third input option allows users to manually input a single gene or multiple genes of interest by pasting the gene list into the web interface.
Users can input gene names separated by commas or on separate lines for quick gene queries.
4.4 Option 4: Genomic coordinates
The fourth input option allows users to upload a set of genomic coordinates representing genomic regions of interest, such as mutation sites or regions with copy number alterations.
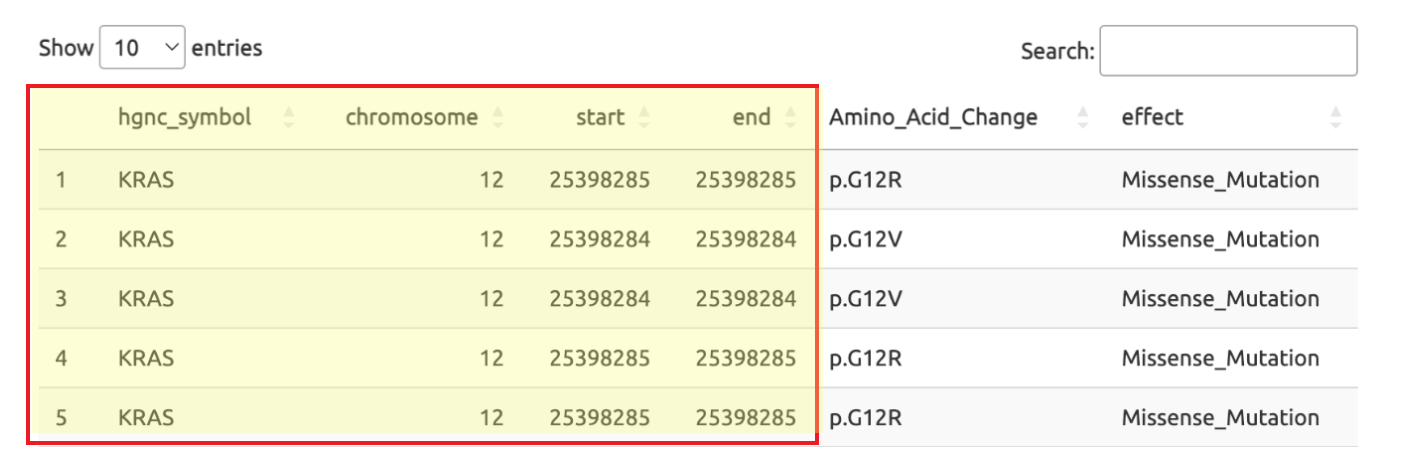
Users must provide four columns of information in their file: hgnc_symbol (if not a gene, this will be the label of the query region), chromosome, start, and end. The coordinates can represent multiple regions within one gene, such as a list of regions with mutations.
Please note that the column names must be the same and additional columns will not affect the reading of the input.Example: