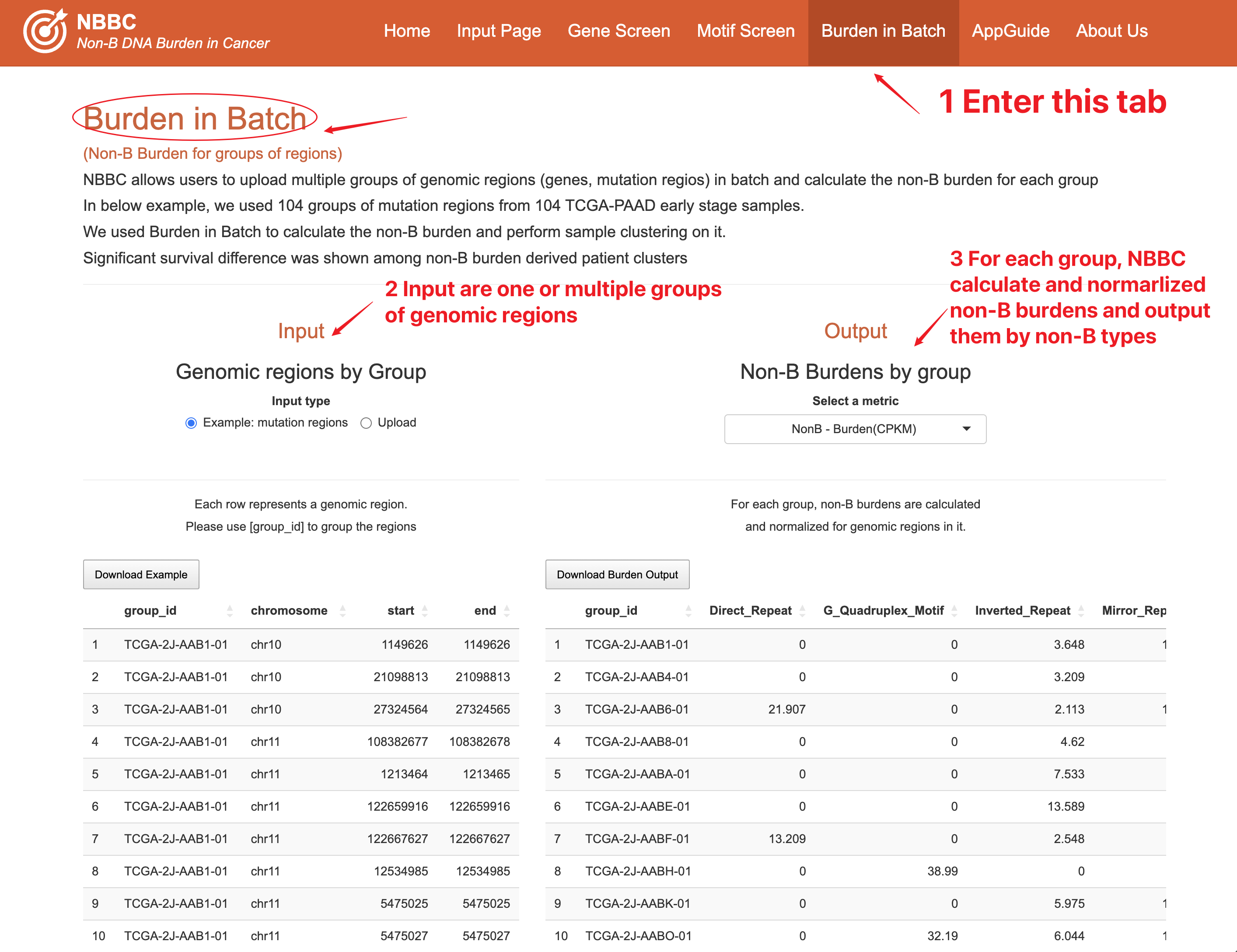
8 More analyses
8.1 Clustering on burdens
Gene/Region Clustering on burdens
Users can choose the columns and rows they want to cluster based on their specific interests, enabling them to identify patterns and relationships between different elements within the heatmap. Additionally, users can select the appropriate scale for the heatmap visualization to ensure optimal visualization of the non-B burden data.

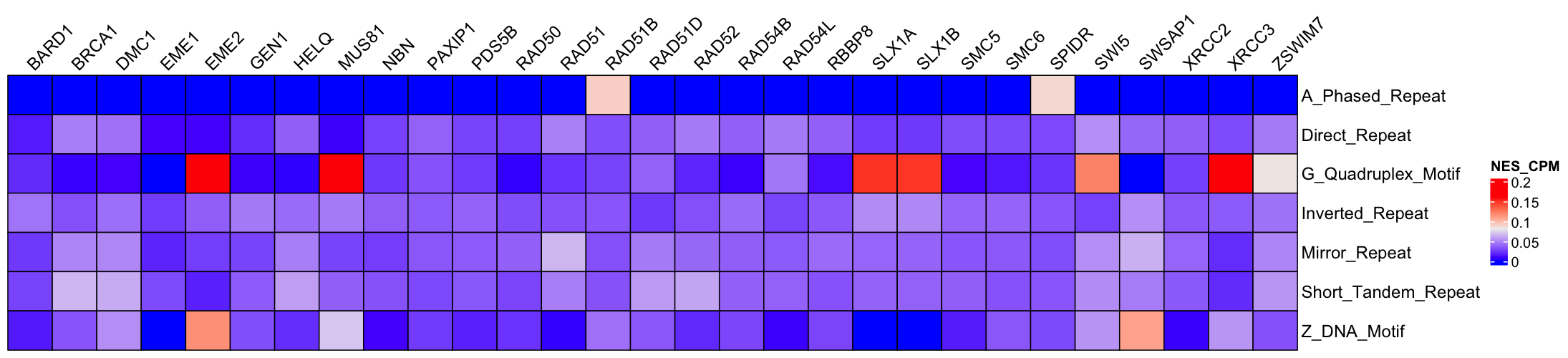
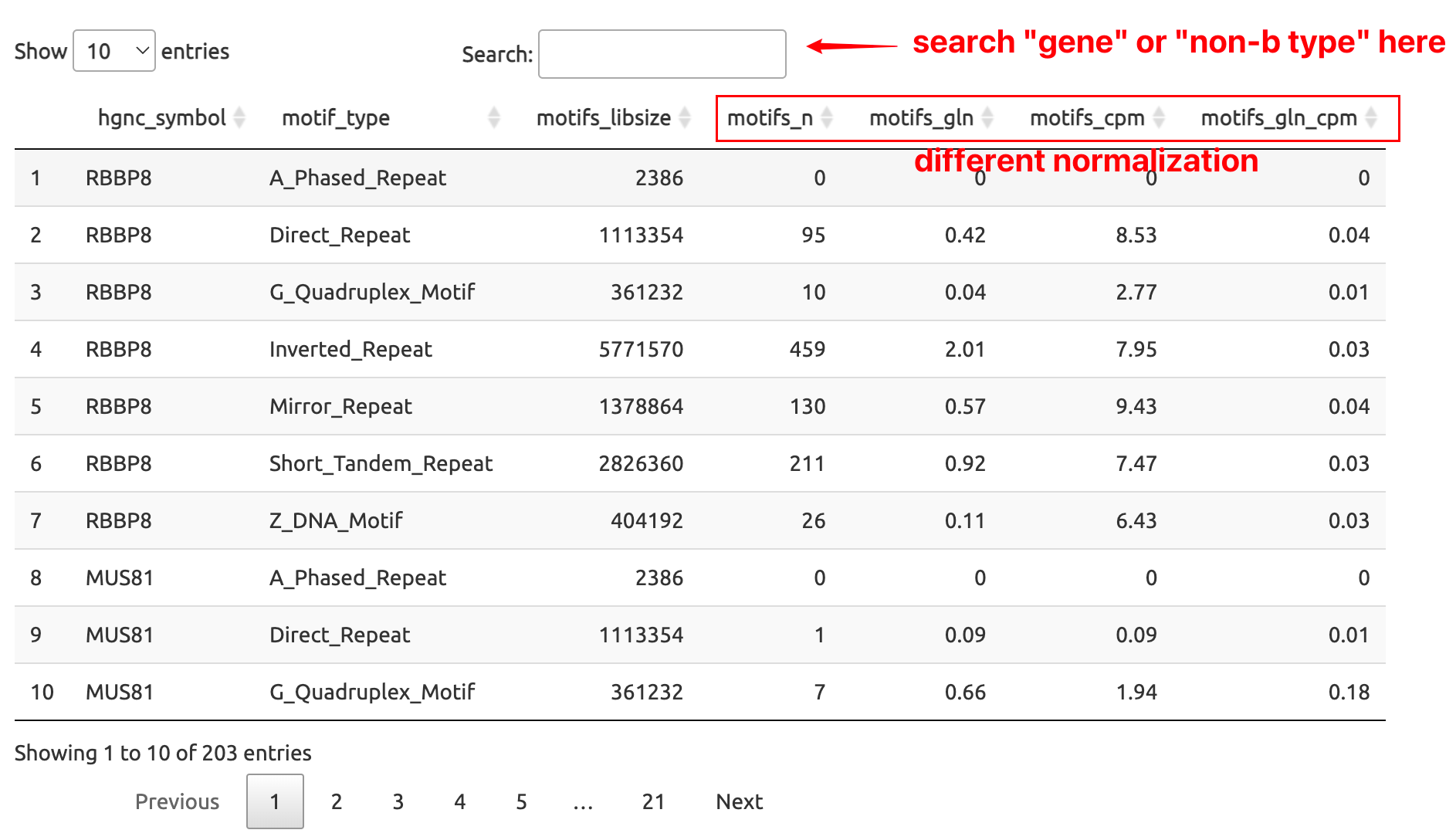
Notes for the names of columns:
motifs_n: Countsmotifs_gln: Normalized by Gene lengthmotifs_cpm: Normalized by Motif Library Sizemotifs_gln_cpm: Normalized by Gene length and Library Size
8.2 Burden in Promoters
For users interested in promoter regions, NBBC provides the option to include a certain region before the start. This feature allows users to include upstream regions of the gene of interest in their analysis, which can be important in identifying potential regulatory elements that may impact gene expression.
This feature within NBBC provides users with a powerful tool for exploring non-B burden in the context of promoter regions and can aid in identifying potential targets for further investigation.
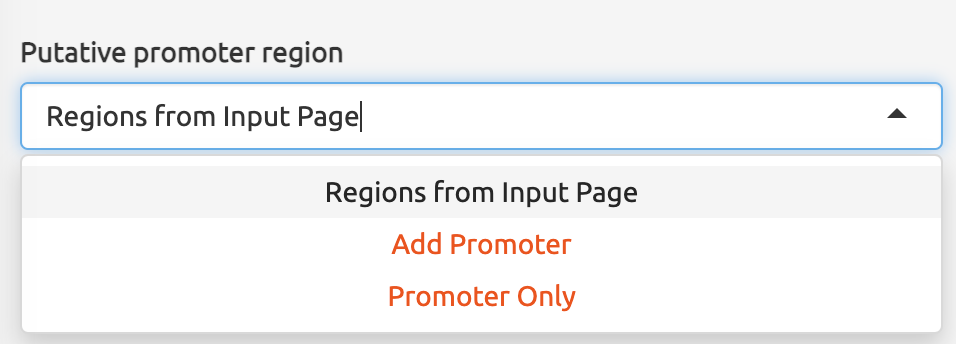
NBBC also provides users with the option to specify the length of putative promoter regions they are interested in. By default, the length is set at 2000 bases, but users can adjust this length to fit their specific research needs.
It is important to note that the option to include putative promoter regions is only applicable for querying non-B motifs in full-length gene regions within NBBC. However, the function will still work for short regions.
For full-length gene regions, the function will extend the query regions for a certain length from the user’s input (e.g. 2000 bases from the “start” column) to include putative promoter regions.
However, for shorter regions, the addition of putative promoter regions may not be relevant, and users should consider this when utilizing the feature within NBBC. This friendly reminder ensures that users have a clear understanding of the functionality of the feature and can utilize it effectively within their research.
8.3 Burden in Batch
In addition to the features previously described, NBBC also offers a ‘burden in batch’ option that allows users to define non-B burden for a set of signatures, such as molecular subtyping, samples, patient-derived models, and more.
This option is particularly useful for generalized queries based on a set of signatures, as it enables users to explore the potential of non-B burden as a marker in downstream analyses and experiments.
The ‘burden in batch’ option can provide valuable insights into the distribution of non-B burden across different signatures, and can help identify potential associations between non-B burden and various biological and clinical factors.
In Case 3 of the NBBC case studies, we demonstrate the application and advantages of using this option in the context of molecular subtyping. Users can refer to the case studies for more information on how to use this feature and its potential benefits.