13 Overview
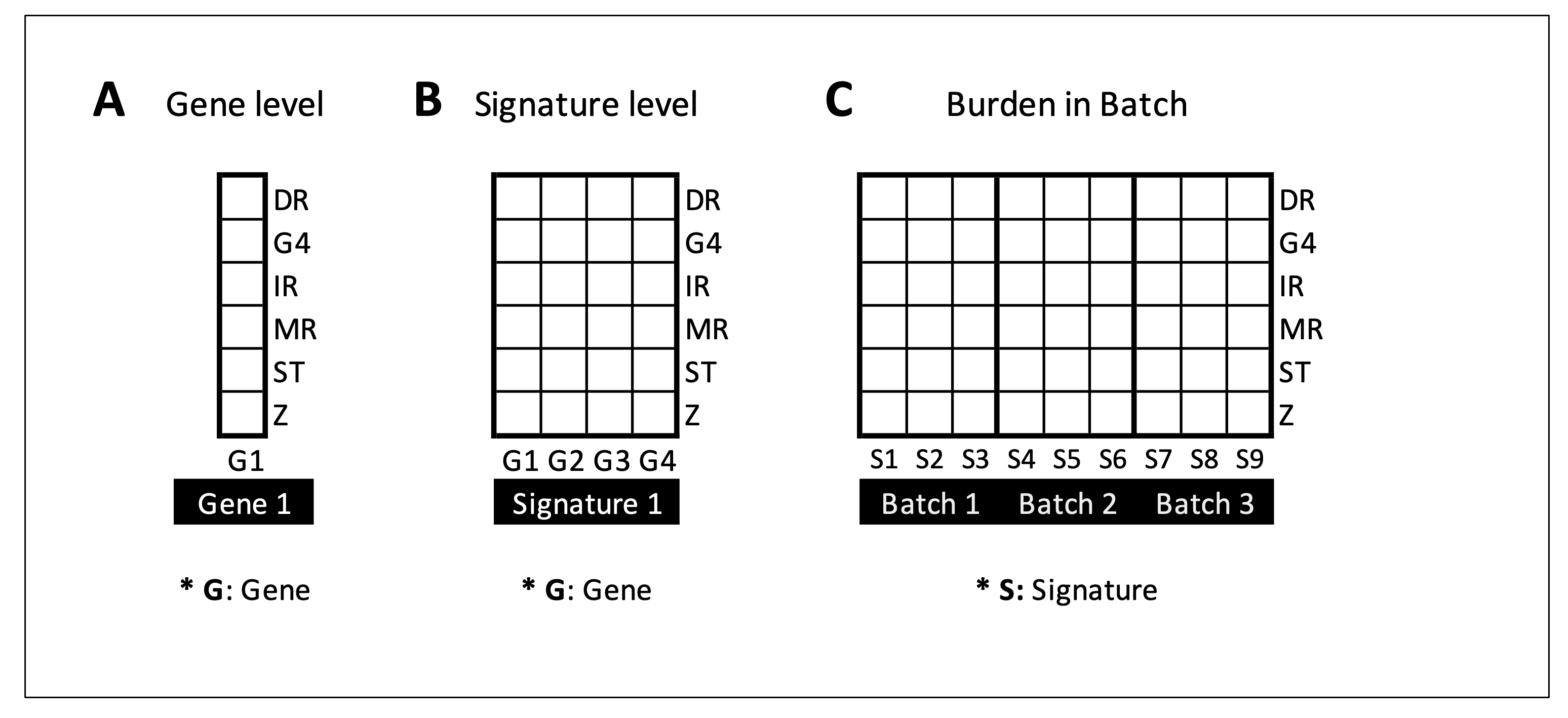
NBBC supports multiple levels of gene and gene regions queries to satisfy potential use cases. The basic input of NBBC includes either gene symbol (gene-or signature-level) or defined genomic regions (site-level), singly or in batch. At the gene-level, users can choose to input a single gene or gene signature.
In case 1, we demonstrate the fundamental query of non-B burden for a single gene and corresponding burden composition among non-B types.
In case 2, we demonstrate user query of a gene signature for non-B burden heterogeneity analyses.
In case 3, users can upload multiple genomic regions, such as at mutation sites, for non-B burden computation. To accomplish this task for multiple regions from multiple samples, the module, “Burden in Batch” was designed. This option is especially useful for generalized queries based on a set of signatures, such as from model systems and molecular subtypes.
We demonstrate in case 3 the application and advantage from use of this option.