6 Input formats
The web server provides four options with which to satisfy users’ input needs. The findTTS workflow takes regions as inputs. The assumed final format is 4 columns for each region (row): name of region, chromosome, start, and end. This input format will find TTSs that are fully contained within these regions and allow for further analysis.
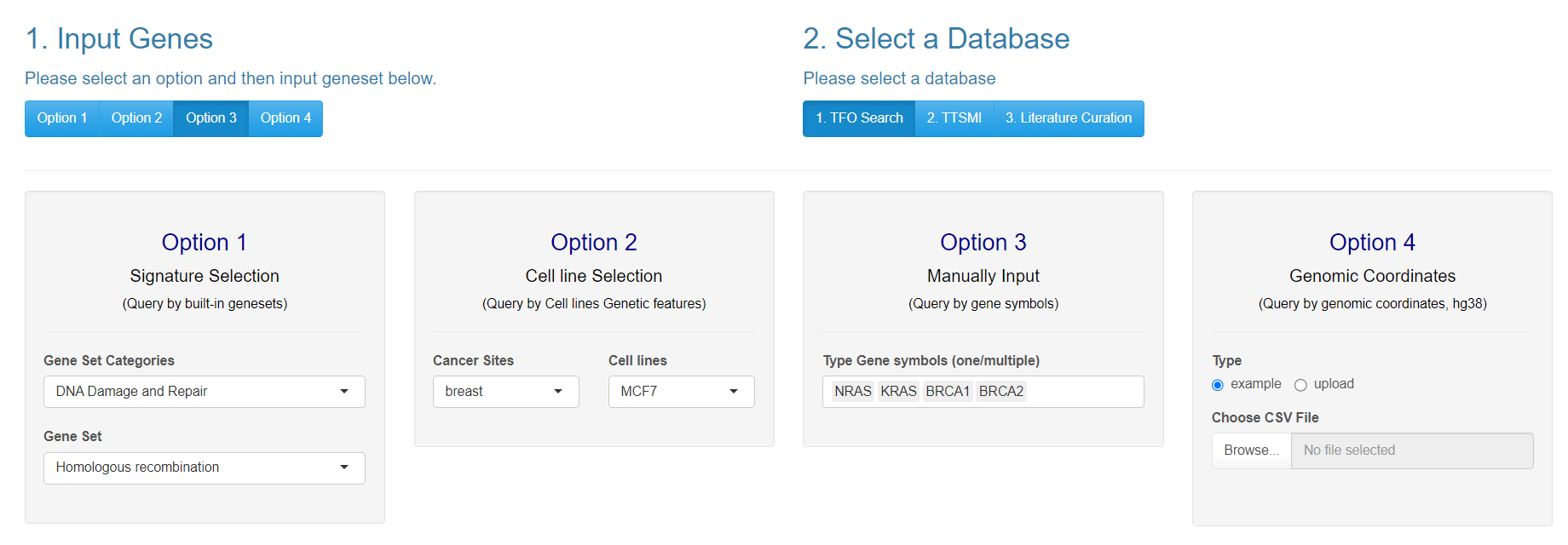
6.1 Option 1: By Cancer Gene Signatures
The first input option includes built-in cancer-related signature gene sets, such as DNA damage repair and response gene pathways, cancer hallmark gene sets, oncogenes, and more. Users can select from these pre-defined sets to perform TTS exploration and analysis for these specific gene sets. These genesets are translated to coordinates based on Gencode v44.
6.2 Option 2: By Cancer Cell Lines
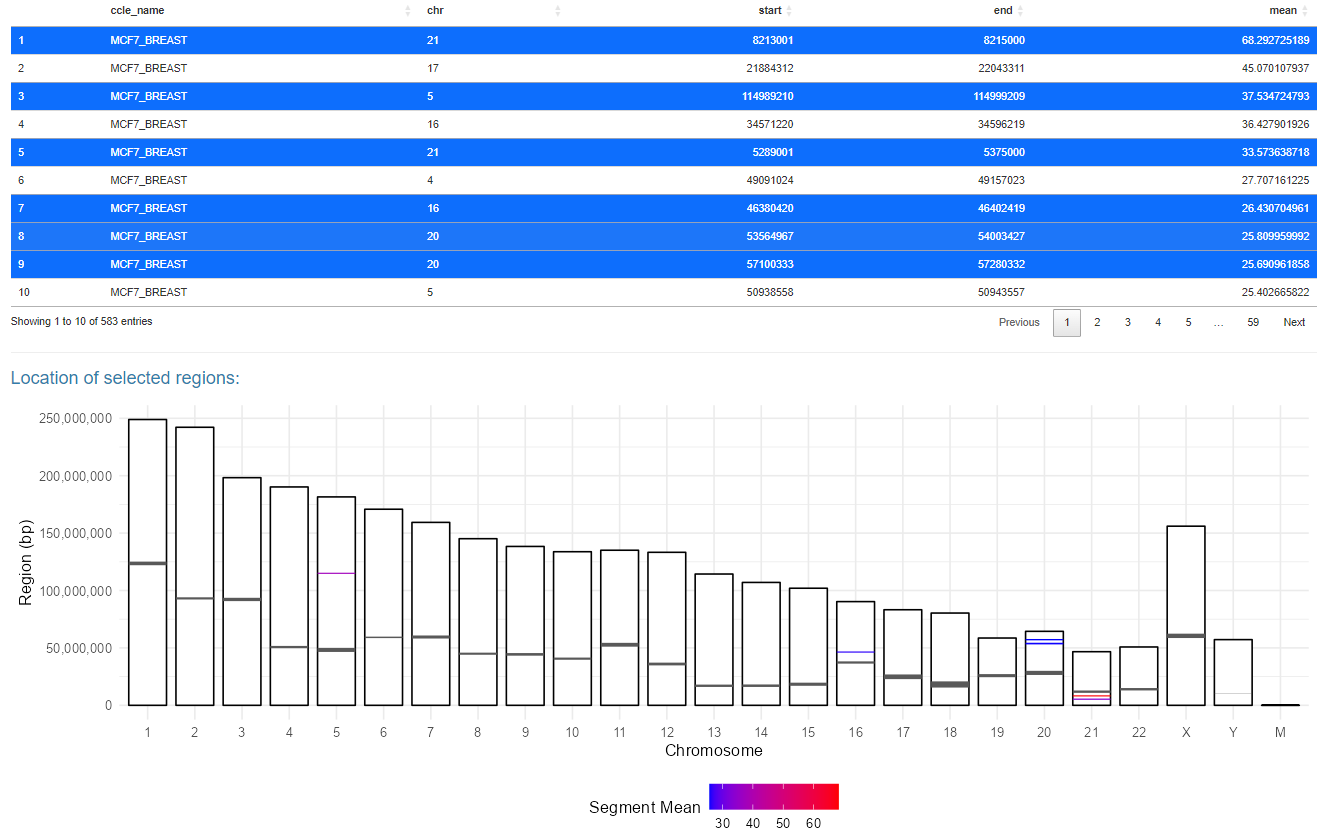
A second input option allows users to select amplified segments from CCLE segments. When using this query option a table will appear with all amplified segments for the selected cell line. A user can select regions from this table by clicking on rows, after clicking the row will change color to indicate the row was selected and the region will appear on the ideogram below.
6.3 Option 3: Custom Gene(set) region(s)
The third input option allows users to select from a drop down list of gene names. Users can input gene symbols that are not in the drop down, but there is no guarantee that they will get correctly converted to coordinates
6.4 Option 4: Custom Genomic coordinate region(s)
The fourth input option allows users to upload a set of genomic coordinates representing genomic regions of interest, such as mutation sites or regions with copy number alterations.
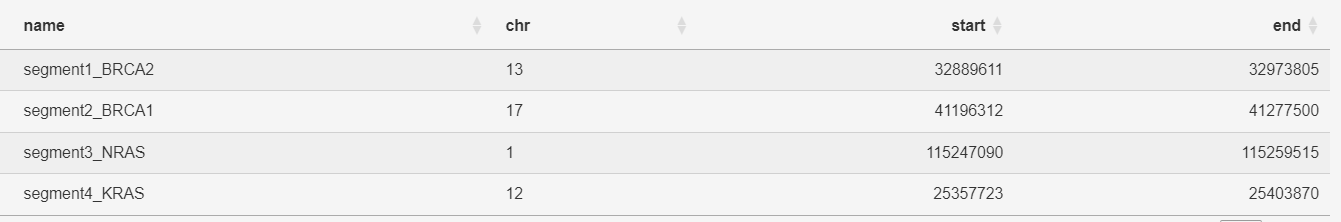
Users must provide four columns of information (with a heading) in their file: name(if not a gene symbol, this will be the label of the query region), chr, start, and end. The coordinates can represent multiple regions within one gene, such as a list of regions with mutations or segments or any other region.
Please note that the column names must be the same and additional columns will not affect the reading of the input. Example: