14 TTS Barcoding
14.1 TTS Barcoding
The TTS Barcoding module of the TTSBBC web server elucidates regions with differences in TTS features. This is especially useful in scenarios where users are interested in differential triplex formation to select regions to target. Multiple metrics for TTS evaluation are provided to cater to the diverse needs of researchers and to support multifaceted investigations of TTSs.
Barcoding enables the categorization of highly specific barcodes for individual TTSs. This feature aids in the precise cataloging and grouping of TTSs based on their features across different genes.
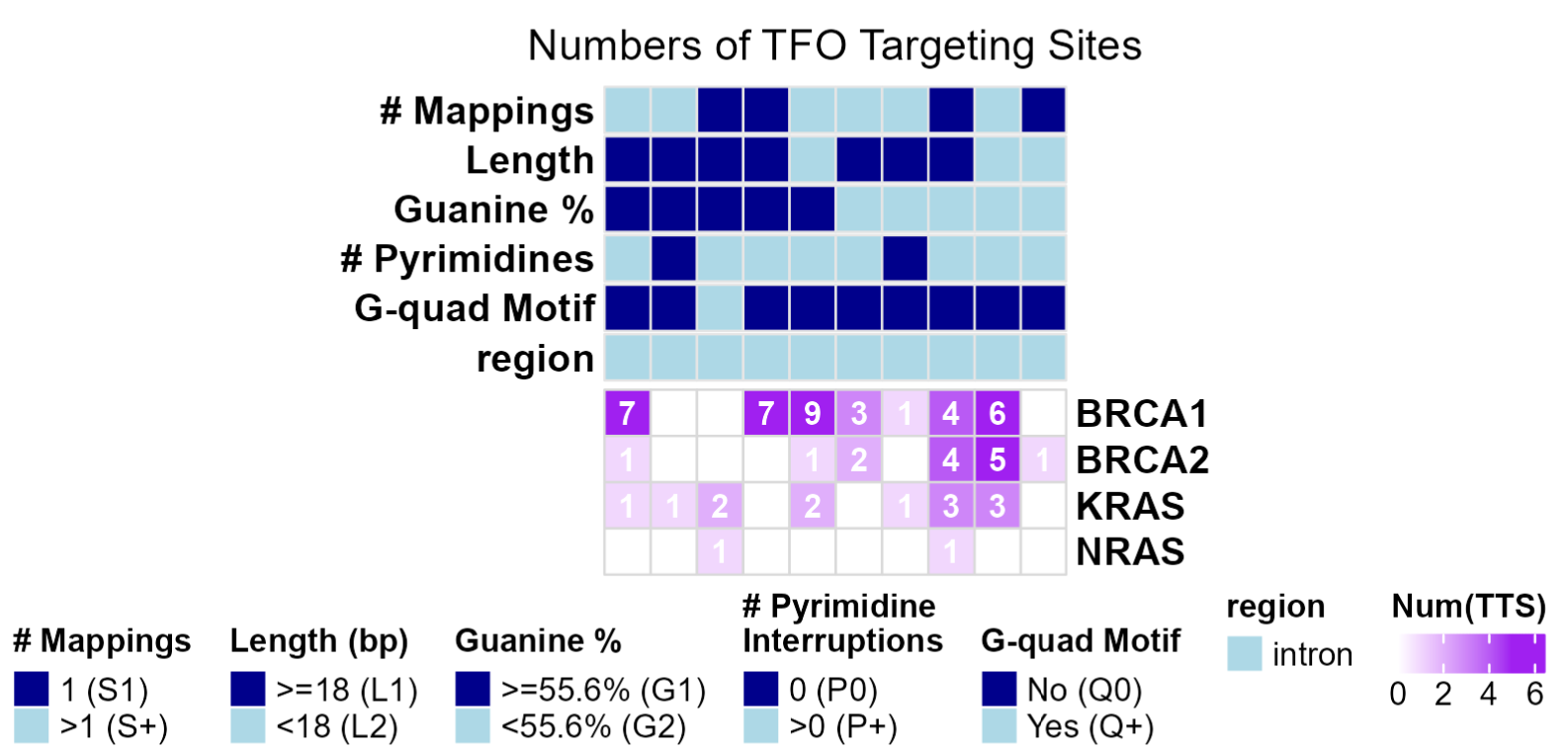
The heatmap assists researchers in understanding the TTS distribution in a more intuitive and user-friendly way, thereby making the process of TTS analysis straightforward and more effective.
Users might notice that there is an increase in the number of TTSs from what they selected in TTS Targeting if and only if there is a TTS that has mapping locations in multiple query regions. For instance if the fake TTS sequence TTAACCGG is in both BRCA1 and BRCA2, then it would be duplicated.
A key for symbols in tables:
- bc_pG = % Guanine barcode
- G1 barcode - high Guanine content, better binding
- G2 barcode - lower Guanine content
- bc_len = Length barcode
- L1 barcode - long length, better binding
- L2 barcode - short length
- bc_spec = Speificity barcode (# Mappings)
- S1 barcode - highly specific barcode
- S+ barcode - low specificity barcode
- bc_num_pyr = Number of Pyrimidines barcode
- P0 barcode - no Pyrimidines barcode, better binding
- P1 barcode - contains at least 1 Pyrimidine
- bc_gquad = G-quadruplex barcode
- Q0 barcode - no G-Quadruplex motif, better binding
- Q1 barcode - contains G-Quadruplex motif